Role of APOBEC proteins in the oncogenesis of HPV viruses
Problem identification
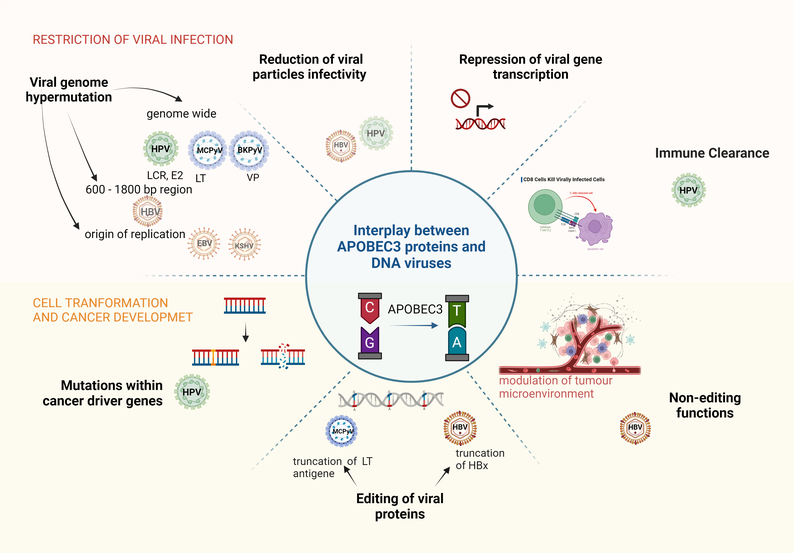
Human papillomaviruses (HPV) are one of the most prevalent sensually transmitted pathogens and are responsible for more than 5% of all human cancers, including cervical cancer and HPV-associated head and neck cancer. APOBEC3 (A3) proteins are cytidine deaminases capable of DNA and RNA editing. DNA editing activity represents an important part of the innate immune response to viral infections. However, this intrinsic host defence mechanism may also be responsible for long-term host DNA hypermutation and cancer development. Recent wide-genome studies have identified APOBECs as the primary source of somatic mutations in HPV-driven cervical cancer, while the molecular mechanisms of oncogenesis remain largely unknown.
Our long-term goal was to elucidate the role of APOBEC proteins in the oncogenesis of HPV viruses. We will approach these processes from various perspectives, including big data biostatistical analyses, genetic studies, and molecular and cell biology experiments.
Results
Objective 1: To analyse genetic and epigenetic changes induced by APOBEC3 expression in cervical cancer samples and HPV model systems.
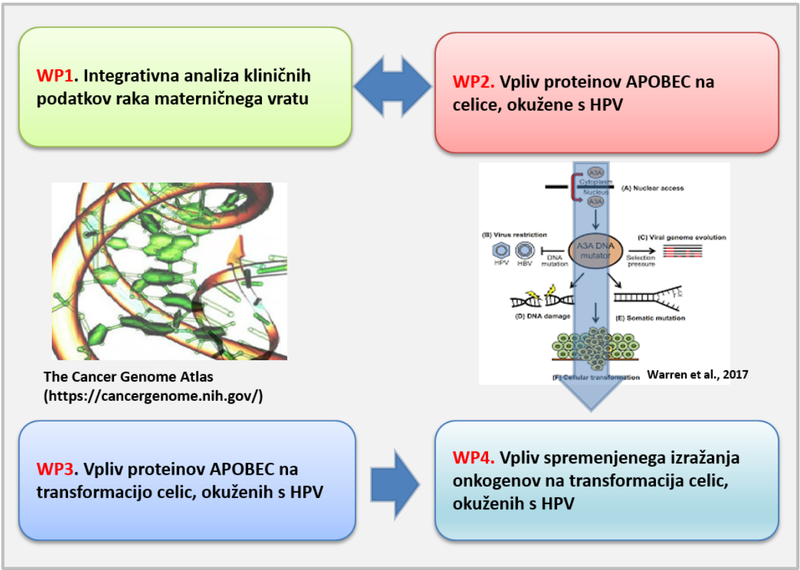
In the first part of the project (WP1), we analysed the expression profiles of patients with cervical cancer (CESC) and head and neck cancer (HNSCC) using publicly available TCGA (The Cancer Genome Atlas) data. This phase resulted in a database of genes whose expression corelates with the expression of APOBEC3A (A3A) and APOBEC3B (A3B) proteins in CESC and HNSCC. The database includes all genes with a correlation greater than 0.3 compared to A3A or A3B expression. For each included gene in HNSCC, data on expression in HPV-positive versus HPV-negative patients and differences in the methylation status of each gene in HPV-positive versus HPV-negative patients are also included. In the second part, publicly available expression data from selected studies were added to the database. The database allows searching by keywords (gene name) and by the degree of correlation with A3A and A3B expression or expression in HPV-positive tissues.
The analysis led to interesting findings regarding the differences between CESC and HNSCC. We demonstrated that the expression of A3A and A3B in CESC positively correlates with a similar set of genes. In HNSCC, however, individual genes correlate with A3A or A3B expression, mostly showing a negative correlation. Genes that positively correlate with A3B expression have higher expression in HPV-positive patients, whereas genes that correlate with A3A expression are more prevalent in HPV-negative HNSCC patients.
In the second part of this segment (WP3), we analysed the transformation potential of HPV host cells HFK and HFK-16 related to the expression of A3A and A3B proteins. We found that silencing the A3A and A3B genes reduces cell metabolism, adhesion, migration, and invasiveness, with the presence of the HPV16 genome (oncogenes HPV E6 and E7) having only a minor effect. We also analysed the expression of HPV E6 and E7 oncoproteins at the transcriptional and protein levels and found that their expression is not associated with the expression of A3A or A3B proteins.
Objective 2: To determine the timeframe and mechanisms of transformation in HPV-infected cells as a result of APOBEC3 protein activity.
To gain insight into the timeline of somatic changes in HPV-infected cells caused by A3 protein activity, we transfected HFK cells with the HPV 16 genome, which initially exists in an episomal form (WP2). During prolonged culture, cells with episomal HPV 16 spontaneously transit to a population containing integrated HPV 16 genomes. HFK cells were successfully transfected with the HPV genome, and viral DNA integration into the host cell genome occurred after approximately 14 months of cell culture. This part of the project was conducted in collaboration with Dr. Vjekoslav Tomaić’s group at the Rudjer Bošković Institute in Zagreb. Transcriptome, methylome, and editome analyses will reveal the genetic and epigenetic changes that occur during the integration of viral DNA into the host cell genome and their approximate timeline.
In the final part of the project (WP4), we used data from WP1 to select specific oncogenes for further analysis. Selection criteria included a high degree of correlation with A3A and/or A3B, differential expression in HPV-positive tissues, and relevant methylation status. Through screening tests, we identified the most promising proteins for biochemical characterization in model systems of different HPV host cells in connection with A3A and A3B.
Conclusions
The research results clearly demonstrated that APOBEC3A and 3B proteins influence the expression of numerous other proteins in tissues infected with HPV viruses. They also have a significant impact on cellular properties crucial for cell transformation, such as proliferation, migration, and invasiveness. These new insights contribute to uncovering the mechanisms of HPV virus oncogenesis, which can indirectly assist the development of clinical and applied research on HPV viruses and other viral infections. This is especially relevant for other small DNA oncogenic viruses, which are also potential targets of APOBEC protein action. This area needs to be investigated promptly to leverage this knowledge for the prevention and treatment of diseases resulting from HPV and other virus infections involving APOBEC proteins. Lastly, the project provided an insight how virus-host interactions drive the co-evolution of viruses and host cells, and how these interactions may lead to unexpected consequences such as cancer development.
Partners in the project: Laboratory for Environmental and Life Sciences – University of Nova Gorica (coordinator), Faculty of Pharmacy – University of Ljubljana, Medical Faculty – University of Ljubljana
Duration: 1 September 2020 – 31 August 2023
Publications:
LOVŠIN, Nika, GANGUPAM, Bhavani, BERGANT MARUŠIČ, Martina. The intricate interplay between APOBEC3 proteins and DNA tumour viruses. Pathogens. 2024, vol. 13, no. 3, art. 187, 19 str., ilustr. ISSN 2076-0817. https://www.mdpi.com/2076-0817/13/3/187, DOI: 10.3390/pathogens13030187. [COBISS.SI-ID 186197507]
SRPČIČ, Anja, LOVŠIN, Nika. Sistem CRISPR/Cas9 in njegova uporaba v genetskem inženirstvu. Farmacevtski vestnik. [Tiskana izd.]. 2023, 74(3), 205-214, https://www.sfd.si/wp-content/uploads/2023/07/srpcic-fv3.pdf [COBISS.SI-ID 160756739]
CARSE, Sinead, BERGANT MARUŠIČ, Martina, SCHÄFER, Georgia. Advances in targeting HPV infection as potential alternative prophylactic means. International journal of molecular sciences. Feb. 2021, vol. 22, iss. 4, str. 1-26, ilustr. ISSN 1422-0067. https://www.mdpi.com/1422-0067/22/4/2201. [COBISS.SI-ID 53587971]
Invited talks/lectures:
BERGANT MARUŠIČ, Martina, LAPENTA, Fabio, LOVŠIN, Nika. APOBEC proteins play an important role in human papillomavirus infection and oncogenesis. Biochemistry and molecular genetics in medicine: Scientific Symposium with International Participation, University of Ljubljana: 6.-7. 7. 2023, Ljubljana: Faculty of Medicine, 2023. Str. 45-46. https://www.mf.uni-lj.si/application/files/6516/8858/1409/Biochemistry_and_Molecular_Genetics_in_Medicine_proceedings.pdf [COBISS.SI-ID 159359747]
LOVŠIN, Nika. Approaches to function analyses (gene overexpression, gene knockout with CRISPR/Cas9 technology): vabljeno predavanje na Gemstone Training School 2022 “Osteoblast cell models and functional evaluation of GWAS hits” Ljubljana, 15.-17. 9. 2022. https://cost-gemstone.eu/wp-content/uploads/Gemstone-workshop-2022-Final-program-.pdf. [COBISS.SI-ID 150760707]
BERGANT MARUŠIČ, Martina. Human papillomaviruses interfere with host cell processes during infection: lecture at ICGEB, Cape Town, South Africa, 5. 7. 2022, on-line. https://www.icgeb.org/martina-bergant-marusic/ [COBISS.SI-ID 114229251]
Presentations at scientific conferences:
LOVŠIN, Nika, LAPENTA, Fabio, PEČAR FONOVIĆ, Urša, JAZBEC, Katerina, MALIČEV, Elvira, BERGANT MARUŠIČ, Martina. Contribution of APOBEC proteins 3A and 3B to the oncogenicity of HPV viruses. 15th Meeting of the Slovenian Biochemical Society with International Participation: book of abstracts: Portorož, 20-23 September 2023. Ljubljana: Slovensko biokemijsko društvo, 2023. Str. 119, p68. [COBISS.SI-ID 165634307]
LAPENTA, Fabio, PAPAROKIDOU, Christina, BERGANT MARUŠIČ, Martina. Correlation of APOBEC3A and APOBEC3B gene expression in HNSCC TCGA study. V: NADIŽAR, Nejc (ur.). 17th CFGBC Symposium: book of abstracts, 6th june 2022. Ljubljana: Faculty of Medicine, 2022. Str. 36. [COBISS.SI-ID 131562499]